T-SNE-Java now have support for Barnes Hut which makes it possible to run the amazing t-SNE on much larger data sets (or much faster on small data sets:) )!
The Barnes Hut version can also be run in parallel! We have seen from 40 % performance improvements on moderate datasets (ca 10 000 samples) to 100 % improvements on larger datasets (MNIST 60000 samples) compared to standard Barnes Hut.
The t-SNE part of running the parallel Barnes Hut t-SNE on MNIST 60000 takes 18.3 minutes on a 2013 Macbook Pro (theta=0.5, perplexity = 50, 1000 iterations)
Both standard and parallel Barnes Hut is of course magnitudes faster than vanilla t-SNE.
Great research by Dr. Maaten!!
Pure Java implementation of Van Der Maaten and Hinton's t-SNE clustering algorithm.
This project is divided into two separate Maven projects, one for the core t-SNE and one for the demos (stand-alone executables that can be run from command line).
If you just want to use TSne as a command line tool, you should use BarnesHutTSneCsv for the Barnes Hut version or TSneCsv for the classic version.
You must then first build and install 'tsne' and 'tsne-demos' (mvn install). Then use the tsne-demos JAR you just build according to the examples below.
You can also download the pre-build binary JAR from the release page.
Examples:
Run TSne on file without headers and no labels.
java -jar target/tsne-demos-2.4.0.jar -nohdr -nolbls src/main/resources/datasets/iris_X.txt Run TSne on CSV file with headers and label column nr. 5.
java -jar target/tsne-demos-2.4.0.jar --lblcolno 5 src/main/resources/datasets/iris.csvRun TSne on file without headers and no labels but supply a separate label file (with the same ordering as the data file).
java -jar target/tsne-demos-2.4.0.jar --nohdr --nolbls --label_file=src/main/resources/datasets/iris_X_labels.txt src/main/resources/datasets/iris_X.txtSame as above but using parallelization.
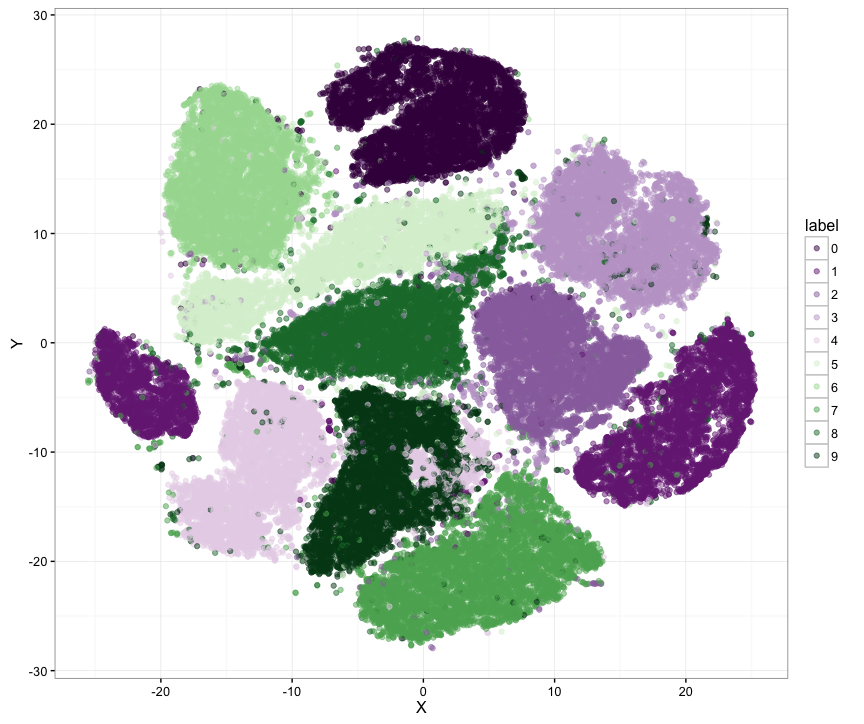
java -jar target/tsne-demos-2.4.0.jar --parallel --nohdr --nolbls --label_file=src/main/resources/datasets/iris_X_labels.txt src/main/resources/datasets/iris_X.txtExample graph of the MNIST data set (60000 samples) generated with Barnes Hut implementation of t-SNE:
For some tips working with t-sne [Klick here] (http://lejon.github.io) or [here] (https://lvdmaaten.github.io/tsne/#faq) (observe that the last link discusses some implementation details of Laurens implementation of t-SNE and not this Java version, but also some general tips and tricks which applies to t-SNE in general) .
import java.io.File;
import com.jujutsu.tsne.barneshut.BHTSne;
import com.jujutsu.tsne.barneshut.BarnesHutTSne;
import com.jujutsu.tsne.barneshut.ParallelBHTsne;
import com.jujutsu.utils.MatrixOps;
import com.jujutsu.utils.MatrixUtils;
public class TSneTest {
public static void main(String [] args) {
int initial_dims = 55;
double perplexity = 20.0;
double [][] X = MatrixUtils.simpleRead2DMatrix(new File("src/main/resources/datasets/mnist2500_X.txt"), " ");
System.out.println(MatrixOps.doubleArrayToPrintString(X, ", ", 50,10));
BarnesHutTSne tsne;
boolean parallel = false;
if(parallel) {
tsne = new ParallelBHTsne();
} else {
tsne = new BHTSne();
}
double [][] Y = tsne.tsne(X, 2, initial_dims, perplexity);
// Plot Y or save Y to file and plot with some other tool such as for instance R
}
}import java.io.File;
import com.jujutsu.tsne.FastTSne;
import com.jujutsu.tsne.TSne;
import com.jujutsu.utils.MatrixOps;
import com.jujutsu.utils.MatrixUtils;
public class TSneTest {
public static void main(String [] args) {
int initial_dims = 55;
double perplexity = 20.0;
double [][] X = MatrixUtils.simpleRead2DMatrix(new File("src/main/resources/datasets/mnist2500_X.txt"), " ");
System.out.println(MatrixOps.doubleArrayToPrintString(X, ", ", 50,10));
TSne tsne = new FastTSne();
double [][] Y = tsne.tsne(X, 2, initial_dims, perplexity);
// Plot Y or save Y to file and plot with some other tool such as for instance R
}
}Demo: 2.4.0 Core: 2.3.0
I'm a very satisfied user of the YourKit profiler. A Great product with great support. It has been sucessfully used for profiling in this project.
YourKit supports open source projects with its full-featured Java Profiler. YourKit, LLC is the creator of YourKit Java Profiler and YourKit .NET Profiler, innovative and intelligent tools for profiling Java and .NET applications.
Enjoy! -Leif