Package implements decision tree and ensemble methods
- ID3 Decision Tree with both numerical and categorical inputs
- Isolation Forest for Anomaly Detection
- Tree Ensembles such as Bagging and Adaboost
Add the following dependency to your POM file:
<dependency>
<groupId>com.github.chen0040</groupId>
<artifactId>java-decision-forest</artifactId>
<version>1.0.3</version>
</dependency>To create and train a ID3 classifier:
ID3 classifier = new ID3();
clasifier.fit(trainingData);The "trainingData" is a data frame which holds data rows with labeled output (Please refers to this link to find out how to store data into a data frame)
To predict using the trained ARTMAP classifier:
String predicted_label = classifier.transform(dataRow);The detail on how to use this can be found in the unit testing codes. Below is a complete sample codes of classifying on the libsvm-formatted heart-scale data:
InputStream inputStream = new FileInputStream("heart_scale");
DataFrame dataFrame = DataQuery.libsvm().from(inputStream).build();
// as the dataFrame obtained thus far has numeric output instead of labeled categorical output, the code below performs the categorical output conversion
dataFrame.unlock();
for(int i=0; i < dataFrame.rowCount(); ++i){
DataRow row = dataFrame.row(i);
row.setCategoricalTargetCell("category-label", "" + row.target());
}
dataFrame.lock();
ID3 classifier = new ID3();
classifier.fit(dataFrame);
for(int i = 0; i < dataFrame.rowCount(); ++i){
DataRow tuple = dataFrame.row(i);
String predicted_label = classifier.transform(tuple);
System.out.println("predicted: "+predicted_label+"\tactual: "+tuple.categoricalTarget());
}To create and train a Bagging ensemble classifier:
Bagging classifier = new Bagging();
clasifier.fit(trainingData);The "trainingData" is a data frame which holds data rows with labeled output (Please refers to this link to find out how to store data into a data frame)
To predict using the trained ARTMAP classifier:
String predicted_label = classifier.transform(dataRow);The detail on how to use this can be found in the unit testing codes. Below is a complete sample codes of classifying on the libsvm-formatted heart-scale data:
InputStream inputStream = new FileInputStream("heart_scale");
DataFrame dataFrame = DataQuery.libsvm().from(inputStream).build();
// as the dataFrame obtained thus far has numeric output instead of labeled categorical output, the code below performs the categorical output conversion
dataFrame.unlock();
for(int i=0; i < dataFrame.rowCount(); ++i){
DataRow row = dataFrame.row(i);
row.setCategoricalTargetCell("category-label", "" + row.target());
}
dataFrame.lock();
Bagging classifier = new Bagging();
classifier.fit(dataFrame);
for(int i = 0; i < dataFrame.rowCount(); ++i){
DataRow tuple = dataFrame.row(i);
String predicted_label = classifier.transform(tuple);
System.out.println("predicted: "+predicted_label+"\tactual: "+tuple.categoricalTarget());
}InputStream irisStream = new FileInputStream("iris.data");
DataFrame irisData = DataQuery.csv(",")
.from(irisStream)
.selectColumn(0).asNumeric().asInput("Sepal Length")
.selectColumn(1).asNumeric().asInput("Sepal Width")
.selectColumn(2).asNumeric().asInput("Petal Length")
.selectColumn(3).asNumeric().asInput("Petal Width")
.selectColumn(4).asCategory().asOutput("Iris Type")
.build();
TupleTwo<DataFrame, DataFrame> parts = irisData.shuffle().split(0.9);
DataFrame trainingData = parts._1();
DataFrame crossValidationData = parts._2();
System.out.println(crossValidationData.head(10));
MultiClassAdaBoost multiClassClassifier = new MultiClassAdaBoost();
multiClassClassifier.fit(trainingData);
ClassifierEvaluator evaluator = new ClassifierEvaluator();
for(int i=0; i < crossValidationData.rowCount(); ++i) {
String predicted = multiClassClassifier.classify(crossValidationData.row(i));
String actual = crossValidationData.row(i).categoricalTarget();
System.out.println("predicted: " + predicted + "\tactual: " + actual);
evaluator.evaluate(actual, predicted);
}
evaluator.report();InputStream irisStream = new FileInputStream("iris.data");
DataFrame irisData = DataQuery.csv(",")
.from(irisStream)
.selectColumn(0).asNumeric().asInput("Sepal Length")
.selectColumn(1).asNumeric().asInput("Sepal Width")
.selectColumn(2).asNumeric().asInput("Petal Length")
.selectColumn(3).asNumeric().asInput("Petal Width")
.selectColumn(4).asCategory().asOutput("Iris Type")
.build();
TupleTwo<DataFrame, DataFrame> parts = irisData.shuffle().split(0.9);
DataFrame trainingData = parts._1();
DataFrame crossValidationData = parts._2();
System.out.println(crossValidationData.head(10));
SAMME multiClassClassifier = new SAMME();
multiClassClassifier.fit(trainingData);
ClassifierEvaluator evaluator = new ClassifierEvaluator();
for(int i=0; i < crossValidationData.rowCount(); ++i) {
String predicted = multiClassClassifier.classify(crossValidationData.row(i));
String actual = crossValidationData.row(i).categoricalTarget();
System.out.println("predicted: " + predicted + "\tactual: " + actual);
evaluator.evaluate(actual, predicted);
}
evaluator.report();To create and train a Bagging ensemble classifier:
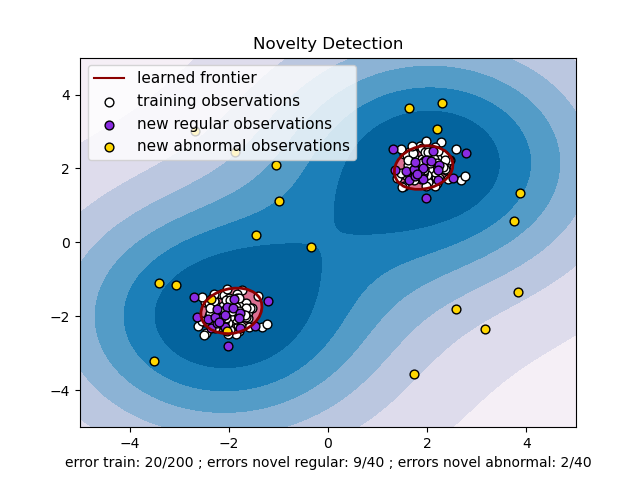
The problem that we will be using as demo is the following anomaly detection problem:
Below is the sample code which illustrates how to use Isolation Forest to detect outliers in the above problem:
DataQuery.DataFrameQueryBuilder schema = DataQuery.blank()
.newInput("c1")
.newInput("c2")
.newOutput("anomaly")
.end();
Sampler.DataSampleBuilder negativeSampler = new Sampler()
.forColumn("c1").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("c2").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("anomaly").generate((name, index) -> 0.0)
.end();
Sampler.DataSampleBuilder positiveSampler = new Sampler()
.forColumn("c1").generate((name, index) -> rand(-4, 4))
.forColumn("c2").generate((name, index) -> rand(-4, 4))
.forColumn("anomaly").generate((name, index) -> 1.0)
.end();
DataFrame data = schema.build();
data = negativeSampler.sample(data, 20);
data = positiveSampler.sample(data, 20);
System.out.println(data.head(10));
IsolationForest method = new IsolationForest();
method.setThreshold(0.38);
DataFrame learnedData = method.fitAndTransform(data);
BinaryClassifierEvaluator evaluator = new BinaryClassifierEvaluator();
for(int i = 0; i < learnedData.rowCount(); ++i){
boolean predicted = learnedData.row(i).categoricalTarget().equals("1");
boolean actual = data.row(i).target() == 1.0;
evaluator.evaluate(actual, predicted);
logger.info("predicted: {}\texpected: {}", predicted, actual);
}
logger.info("summary: {}", evaluator.getSummary());