-
Notifications
You must be signed in to change notification settings - Fork 0
Home
CTAT-LR-Fusion is a component of the Trinity Cancer Transcriptome Analysis Toolkit (CTAT) used for detecting fusion transcripts from long-read transcriptome sequencing data, including PacBio Iso-seq and Oxford Nanopore Technology sequenced transcriptomes. If matched Illumina RNA-seq data are available, these can be leveraged as well for additional exploration and quantification of fusions initially detected via long reads.
CTAT-LR-Fusion was developed in the Broad Institute's Methods Development Laboratory (MDL) for characterizing long read transcriptome sequences such as derived from MAS-seq.
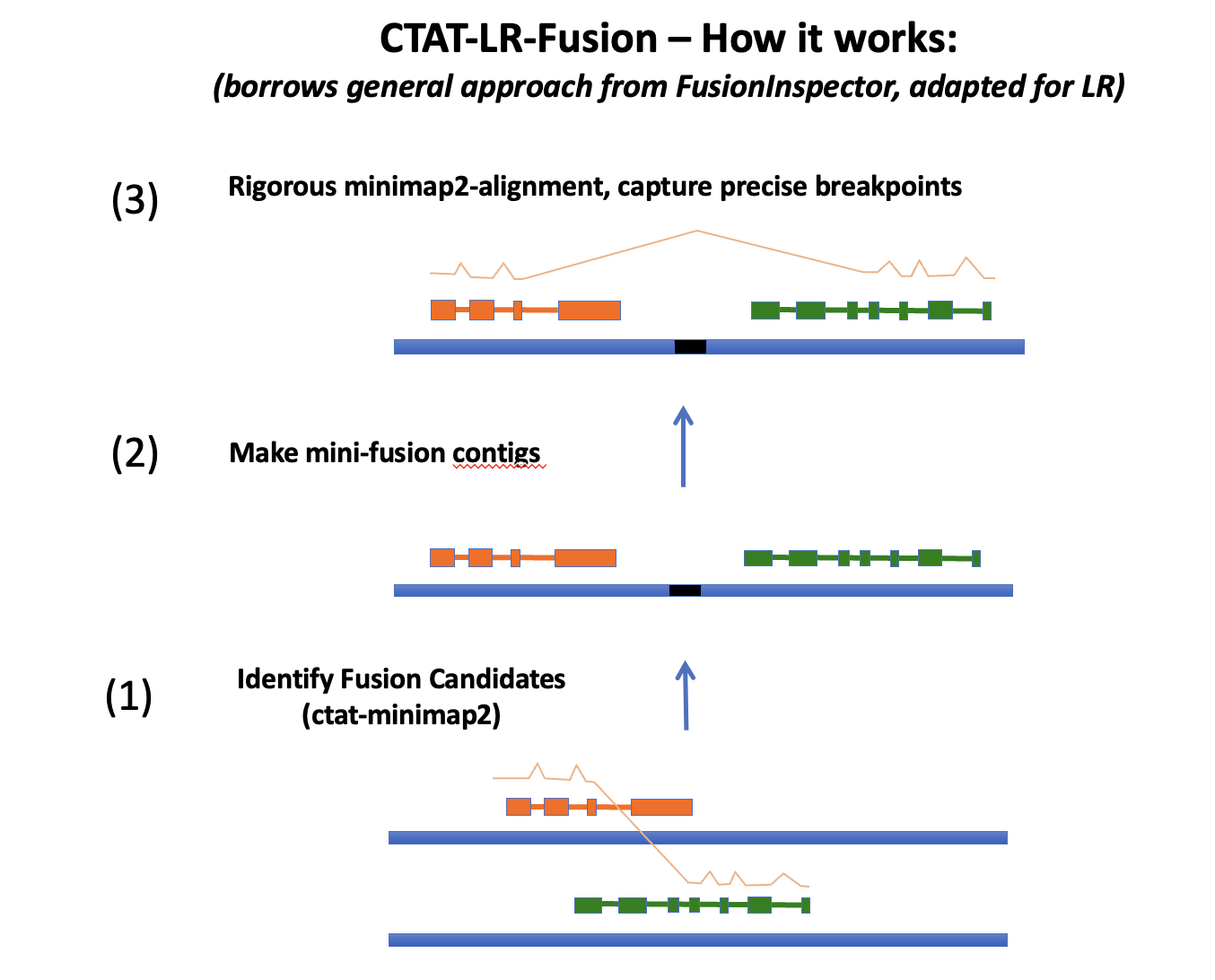
CTAT-LR-fusion operates in three main steps:
-
Fusion candidates are initially identified based on long read alignments using ctat-minimap2, a modified version of minimap2 that focuses on identifying likely chimeric long reads rather than providing high quality alignments for all input reads. The chimeric-read-only search speeds up the initial minimap2 search phase.
-
The chimeric read alignments are screened based on read and genome alignment positions to define a list of fusion candidates. For each fusion candidate, a model of the ordered and oriented fusion pair is constructed - borrowing the approach from our FusionInspector software.
-
The candidate chimeric reads are realigned to a database of these fusion contigs and each fusion pair is scored for read support according to read alignment breakpoints (aka. fusion transcript breakpoints). If matched Illumina short reads are available, these are separately aligned to these fusion contigs using FusionInpsector and the results are integrated into the final ctat-LR-fusion report with fusion variant expression estimates from both short and long reads, respectively.
Sometimes the short reads provide evidence for alternatively spliced fusion isoforms for which long reads weren't captured, or vice-versa. These cases can be easily identified in the ctat-LR-fusion report.
Docker and Singularity images are available and recommended.
If you would prefer to install from source code, download the latest 'FULL' release tarball from the CTAT-LR-Fusion release site. Unpack it, and run 'make' in the base installation directory.
There are likely other dependencies that you may require. The full installation for a full stack of dependencies is shown in this Dockerfile. You can probably just get away with the following if you're only running long reads through:
pip install pandas igv-reports pysam
The CTAT genome lib is the same used for other CTAT tools and can be downloaded from https://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB/. The ctat genome lib software compatibility matrix indicates the version of STAR to use if you have companion Illumina short reads.
The ctat-LR-fusion software comes with a customized version of minimap2 named ctat-minimap2, and CTAT-LR-Fusion requires a minimap2 index of the reference genome. To build this, initially run ctat-LR-fusion like so:
ctat-LR-fusion -T long_reads.fastq.gz \
--genome_lib_dir /path/to/ctat_genome_lib_build_dir \
--prep_reference --CPU 4
and it will first build the minimap2 genome index before running ctat-LR-fusion to find fusion transcripts.
If you run with --prep_reference_only, it will stop after building the index.
For future runs, drop the --prep_reference argument, as the index only needs to be built once. If you forget, no worries. It'll only build it once anyway.
Once you have the ctat genome lib installed and configured as above.
For long reads, you need either a FASTA or FASTQ formatted file. Then, run ctat-LR-fusion like so:
ctat-LR-fusion -T long_reads.fastq.gz \
--genome_lib_dir /path/to/ctat_genome_lib_build_dir \
--CPU 4 \
--vis
If you have the ctat genome lib dir set up as an environmental variable CTAT_GENOME_LIB, then you don't need to specify --genome_lib_dir, and only need to specify -T for the long reads.
If you have reads that align to the reference genome with <90% sequence identity, adjust the --min_per_id parameter (default: 90) accordingly.
If you additionally have Illumina RNA-seq for the sample, you can include that as well like so:
ctat-LR-fusion -T long_reads.fastq.gz \
--genome_lib_dir /path/to/ctat_genome_lib_build_dir \
--left_fq illumina_reads_1.fq \
--right_fq illumina_reads_2.fq \
--CPU 4 \
--vis
ctat-LR-fusion does not find additional fusions based on short reads... it will only additionally examine short read support for those fusion gene pairs initially detected via long read sequences. However, it will identify fusion splicing isoforms that are uniquely supported by Illumina short read data.
See the full usage info (via --help or no parameters) for additional options and configurations.
The output files consist of the following:
-
ctat-LR-fusion.fusion_predictions.tsv : the final fusion predictions including names for the evidence reads. See the .abridged version for simpler output lacking the read names.
-
ctat-LR-fusion.fusion_inspector_web.html : the results in an interactive igv-reports for exploring the evidence supporting each fusion. Requires the --vis command line argument to ctat-LR-fusion.
A preliminary list of fusions before any filtering is performed to generate the final list is provided as file 'ctat-LR-fusion.fusion_predictions.preliminary.tsv'. This is useful for additional exploration and for troubleshooting purposes.
A screenshot of the interactive fusion html view is shown below:
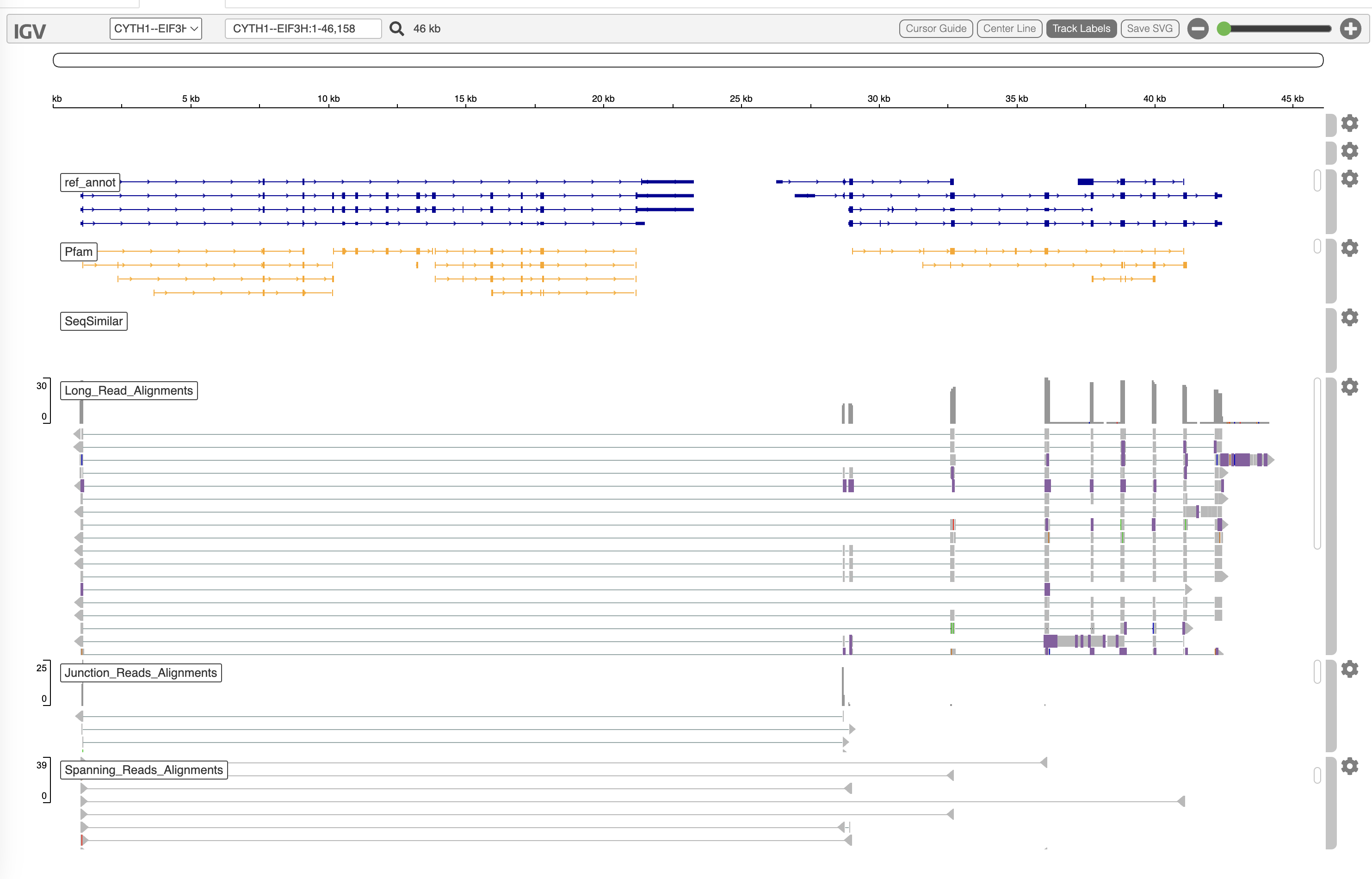
In the image above, we have PacBio Iso-seq reads supporting the fusion, and below Illumina junction reads and spanning fragments that also support this fusion. If you only have long reads, the Illumina tiers will simply be empty. The different fusion breakpoints are evidence of alternatively spliced fusion transcripts from within the single sample.
Before running single cell RNA-seq through CTAT-Mutations, the names of the reads should be encoded with cell barcode and UMI information in the following format:
cellbarcode^UMI^read_name
If you have 10xGenomics reads in a ubam format, you can convert to fastq format with the above read name encoding using this script: 10x_ubam_to_fastq.py
The fusion-to-cell mapping information can be derived from the ctat-LR-fusion output file 'ctat-LR-fusion.fusion_predictions.tsv' using this script: cell_to_fusion_mappings.Rscript, generating a report like so:
FusionName LeftGene LeftBreakpoint RightGene RightBreakpoint SpliceType cb umi readname
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CGAGCCATCTACTATC CTACGGCGGC m64020e_210506_132139/1068814535003/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CAGCCGACAGGACCCT GATTGGTCAA m64020e_210506_132139/1162007282005/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CTACCCATCCAAATGC TCTACGGCGG m64020e_210506_132139/1130810518002/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CGACTTCTCCAAGCCG TGTTGTCTAC m64020e_210506_132139/1109709689001/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CTCTAATTCTCGTATT TTGTTTCGTT m64020e_210506_132139/1016517571004/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE CCCTCCTCAGCTTCGG TACGACCGCA m64020e_210506_132139/1116984222006/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE TTGACTTAGGGTATCG GGTCGGGAGT m64020e_210506_132139/1114230755010/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE TGGTTAGAGACCCACC TTTCCTCCGA m64020e_210506_132139/1009833045005/ccs
NUTM2A-AS1--RP11-203L2.4 NUTM2A-AS1 chr10:87326630:- RP11-203L2.4 chr9:68822648:- ONLY_REF_SPLICE TGGGCGTTCACTGGGC ACATGTATAC m64020e_210506_132139/1164366124008/ccs
...

Contact us via our google group: https://groups.google.com/forum/#!forum/trinity_ctat_users