-
Notifications
You must be signed in to change notification settings - Fork 2
/
README.rmd
125 lines (75 loc) · 4.76 KB
/
README.rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
---
output: github_document
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
```{r, echo = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/"
)
```
# tidyNano <img src="https://raw.githubusercontent.com/nguyens7/tidyNano/master/man/figures/tidyNano.png" align="right" alt="" width="120" />
tidyNano is an R package that imports raw NanoSight data and provides a framework to clean, analyze, and visualize nanoparticle analysis data. You can browse the source code on [GitHub](https://github.com/nguyens7/tidyNano).
Check out our manuscript in [PLOS ONE](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0218270)!
<img src="https://raw.githubusercontent.com/nguyens7/tidyNano/master/man/figures/tidyNano_PLOS_ONE_site_image.png" align="center" width = "600"/>
A representative video of NTA data of murine plasma exosomes from our paper.

# News (7/2020)
Added more [reprexes](https://www.tidyverse.org/help/) in the README to assist in importing data.
Fixed `custom_name` argument in `nanoimport()` function and `nanocombine()` function.
`nanoimport()` has just been updated to be more flexible and works with NTA versions 3.0 - 3.4! The function should be able to detect and import your data.
`nanolyze()` and `nanocount()` both will `ungroup` data.
**Nomenclature**
We recommend naming your files in `snake_case` so it's conducive for processing with the `nanotidy()` function. With each sample we recommend adding important information so you can parse it out during analysis, be sure to influde the dilution value in the sample name if you do not enter it during the dilution entry box during acquisition. For example you can name your files using the general formula `SampleID_Treatment_1500_`. Be sure to leave a `_` at the end so the NTA software will append your files with a numerical count (00,01,02...) which will serve as way to track your technical replicates.
If you already collected you data and don't have your dilution in your sample name you can simply add the dilution when you use `nanoimport()`. Simply add the dilution value in the `custom_name` argument. If you did label your samples and enter in the dilution in the NTA software during acquisition you can make `auto_name == TRUE` in `nanoimport()`.
# Overview
<img src="https://raw.githubusercontent.com/nguyens7/tidyNano/master/man/figures/tidyNano_Schema.png" align="center" width = "600"/>
tidyNano functions (Purple) allow for easily extracting and converting raw NTA count data into a tidy dataframe that is suitable for analysis using dplyr and ggplot2. tidyNano also provides a interactive shiny application (shinySIGHT) for visualizing data.
<img src="https://raw.githubusercontent.com/nguyens7/tidyNano/master/man/figures/tidyNano_workflow.png" align="center" width = "400"/>
tidyNano also includes a Shiny app named shinySIGHT for interactive visualization of NTA data.
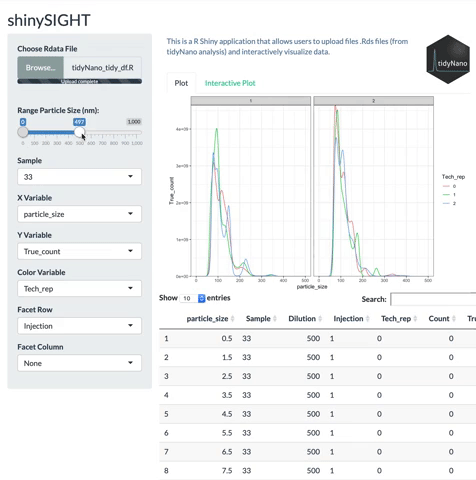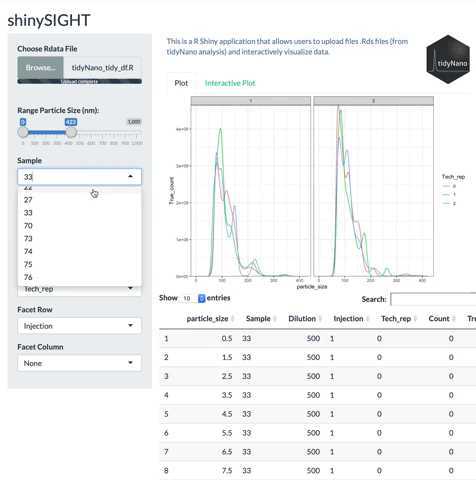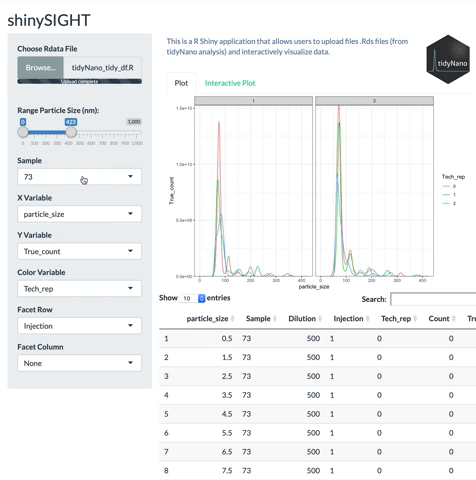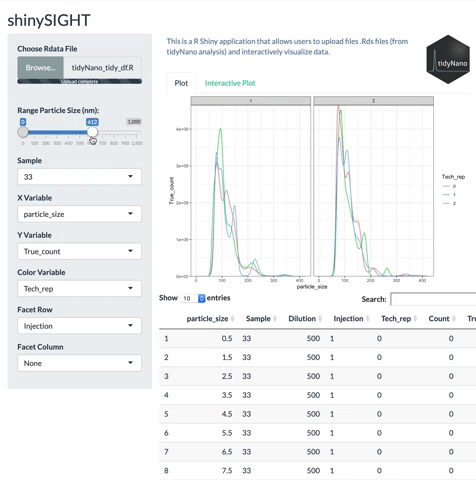
# Installation
The latest development version can be installed from github:
```{r message=FALSE, warning=FALSE}
# install.packages("devtools")
# devtools::install_github("nguyens7/tidyNano")
```
# tidyNano Example
----------------
```{r message=FALSE, warning=FALSE}
library(tidyNano)
library(tidyverse)
file <- system.file("extdata", "beads.csv", package = "tidyNano")
data <- nanoimport(file)
head(data)
```
`nanoimport` is a function that extracts the particle data from raw a nanosight .csv file and creates a dataframe that is suitable for cleaning within R. *Note:* This assumes you added the dilution factor when you named your samples during acquistion.
```{r}
data <- nanoimport(file)
head(data)
```
### nanoimport without dilution factor in the sample name
If you didn't include your dilution factor in the sample name you can use the argument `auto_name == TRUE` within the `nanoimport()` function.
```{r}
file2 <- system.file("extdata", "beads2.csv", package = "tidyNano")
data2 <- nanoimport(file2, auto_name = TRUE)
head(data2)
```
You can even add a custom name to append extra information to your sample columns using the `custom_name()` argument.
```{r}
custom_name_data2 <- nanoimport(file2, auto_name = TRUE, custom_name = "YourLabelHere")
head(custom_name_data2)
```
```{r}
tidy_data <- data %>%
nanotidy(sep_var = c("Sample", "Dilution","Filter","Injection","Tech_rep"))
head(tidy_data)
```
```{r eval=FALSE}
tidy_data %>%
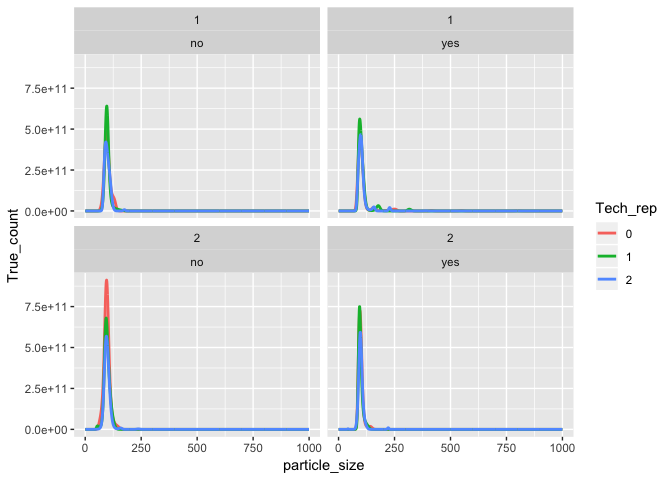
ggplot(aes(x = particle_size, y = True_count, color = Tech_rep)) +
geom_line(size = 1) +
facet_wrap(Injection ~ Filter)
```